This article was presented at a workshop in the 2022 Annual Meeting of the American Scientific Affiliation.
We acknowedge support by a visiting professorship funded by the John Templeton Foundation and hosted by the Southeastern Baptist Theological Seminary in 2021-2022. The author is grateful to several scholars for reading and providing feedback on a draft of this manuscript: John Wood, Ken Keathley, Michael Murray, William Lane Craig, Ken Turner, Andrew Loke, Jeff Hardin, Jeff Schloss, Greg Cootsona, Gerry McDermott, AJ Roberts, Glenn Branch, Ted Davis, and Daniel Ang.
Deborah Haarsma, Jim Stump, Loren Haarsma, Ard Louis, and Dennis Venema from BioLogos were also presented a draft of this manuscript, but they did not provide a response.
The science of genetic bottlenecks is known to many, but often misunderstood. A 2010 article laid out a specific genetic argument against Adam and Eve.1 This argument was expanded substantially outside peer-review, leading to coverage in Christianity Today2 and coverage by National Public Radio (NPR)3 in 2011. This genetic argument, summarized, was that
- Genetic evidence—primarily linkage disequilibrium (LD) and incomplete lineage disequilibrium (ILS) estimates of effective population size (
)—demonstrates that the human lineage was never less than 10,000 individuals going back 18 million years, long before we could remotely be called “human.”4 - Therefore, we do not all descend from Adam and Eve; “That would be against all the genomic evidence that we’ve assembled over the last 20 years.”5
This is a theologically potent argument because it appears to disprove the doctrine of monogenesis.
Beyond the scope of this review, the second point does not follow from the first. Universal ancestry does not require a genetic bottleneck and is entirely consistent with large population sizes.6 This review, however, focuses on the first point, which was also incorrect.
The goal of this review is to explain what the evidence in genetics tells us about single-couple bottlenecks, clarifying each line of evidence. The narrow focus is single-couple “genetic bottlenecks,” and the findings here do not generalize to most models of Adam and Eve. This is a model in which humanity arises from a single couple with normal biology, and there is absolutely and precisely zero interbreeding between their lineage and others. Findings about genetic bottlenecks do not apply to any model that (1) allows any interbreeding at all between Adam and Eve’s lineage and others, or (2) posits exotic biology in Adam and Eve.
There are three key corrections to the first point.7
The genetic evidence only rules out a single-couple bottleneck (2, not 10,000) going back about 500,000 years (not 18 million).
LD, ILS, and
are irrelevant lines of evidence, that do not rule out a single-couple bottlenck in any relevant time scale. Instead, the primary line of evidence establishing the revised conclusion, the most recent time to 4-alleles (TMR4A), was first reported in 2017.
In 2020, the BioLogos organization acknowledged that they did not accurately represent the scientific evidence. By 2021, the organization deleted or substantially edited all the articles related to and supporting the 2011 coverage by Christianity Today. These corrections, however, were not transparent or complete. Consequently, recent academic literature still cites several articles with substantial errors, including articles deleted by the organization.
Rigorous scholarship on any topic engages prior work in detail. The BioLogos organization published more scholarship about genetic bottlenecks than any other group aligned with mainstream science. For this reason, this review engages the organization past and present position, with attention to where corrections have and have not been made. Clarifying the organizations editorial practices, in focused cases, is particularly important where corrections were incomplete, and where mistakes were reasserted.
Going forward, this review is entirely aligned with mainstream science, including evolutionary science. We grant from the outset that the earth looks very old, because it is old. The genetic evidence indicates humans also share common ancestors with the great apes: chimpanzees, bonobos, orangutans, and gorillas. This aspect of BioLogos’s work was not found in fault.
Genetic bottlenecks, for better or worse, are likely to be discussed for some time to come. If we are to discuss bottlenecks, the scientific claims we make about them should be accurate, and not over-extend to inaccurate claims about models that do not require bottlenecks.
Post-Publication Review of Adam and the Genome
In 2017, Dennis Venema and Scott McKnight published Adam and the Genome. Chapter 3 of the book focused on the argument for large ancestral population size as evidence against Adam and Eve. As stated in the book,
All methods employed to date agree that the human lineage has not dipped below several thousand individuals for the last 3 million years or more – long before our lineage was even remotely called “human.”8
In early 2017, a British evolutionary biologist, Richard Buggs, raised specific concerns about the scientific argument in Chapter 3 of Adam and the Genome.9 This chapter of the book focused on the argument for large ancestral population size as evidence against Adam and Eve. In 2017, BioLogos published two articles by Venema defending this scientific argument from Bugg’s criticism.10 In the forum discussion of the first article, a 514-page exchanged unfolded between several scientists, including Buggs and Venema, but also Stephen Schaffner, Ann Gauger, and S. Joshua Swamidass.11 Several others were watching closely from the side-lines, including philosopher William Lane Craig, biologist AJ Roberts from Reasons to Believe (RTB), and several secular scientists at Peaceful Science.
In his initial response, Dennis Venema began with a walk-back, stating that he only meant “human” to refer to our species, Homo sapiens.12 This is a conclusion-altering concession—inconsistent with his prior work and the text of Adam and the Genome itself—because this revision allows for a bottleneck at the headwaters of when most scientists teach our lineage is called “human.”13
Venema also clarified he was only interested in models of a single couple “genetic bottleneck,” which he meant to exclude even a single interbreeding event between Adam and Eve’s lineage and others.
He thought the evidence ruled out a bottleneck going back 18 million years but was most certain a bottleneck would be ruled out at the origin of Homo sapiens, about 200,000 years ago. But if the focus is on our species, this corresponds to a model which excludes all interbreeding between Homo sapiens and other hominids. Most old earth creationists and young earth creationists agree Neanderthals and Homo sapiens interbreed.14 When asked for any evidence that there were never just two Homo sapiens amidst a larger population, Venema could not provide any. So, it must be emphasized, Venema’s revised conclusion is almost entirely irrelevant to the theological conversation on origins.
Charitably biasing the discussion towards Venema, we focused on determining which lines of evidence (beyond interbreeding) ruled out a bottleneck of two individuals (not several thousand) in at least the last ~200,000 (not 3 million) years, a time that is long after (not long before) archaic “humans” arise about 2 million years ago.
Even under these most favorable of terms, the primary evidence in Adam and the Genome did not support the revised conclusion. Several lines of evidence were discussed in detail (Figure 1), but they were all irrelevant, invalid, disputed, or did not cover the relevant time periods. Eventually, another scientist produced a new line of evidence that could estimate the minimum possible population size, the time to most recent 4 alleles (TMR4A).
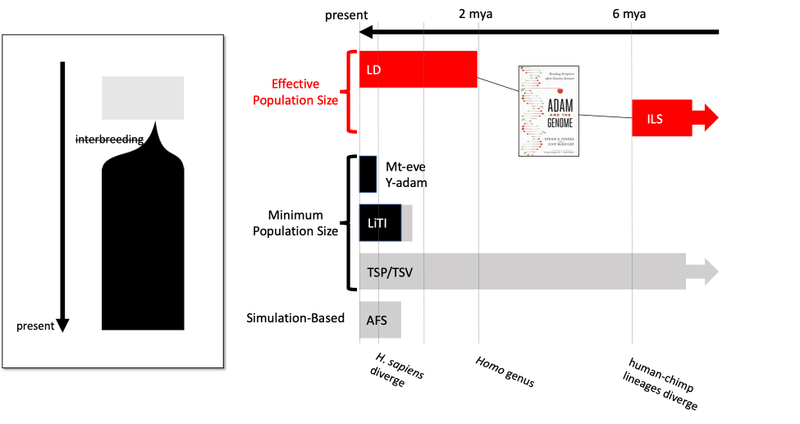
Irrelevant Lines of Evidence
Since 2010, Venema and the BioLogos organization presented two primary lines of evidence to demonstrate large population sizes in the past. Both these lines of evidence failed to establish their claims.
Linkage Disequilibrium and Effective Population Size
The first line of evidence was linkage disequilibrium, which provides a way of estimating “effective population size” (
- Linkage disequilibrium (and other methods) estimates our “effective population size” (
) in the past.15 estimates are always in the thousands or more. is the minimum population size consistent with the data.16 - Therefore, the human lineage “was never less than” several thousand.
There are also several variations of this basic argument that substitute LD with other methods of estimating
Still, in post-publication review, the argument quickly collapsed. First, Venema had claimed that
Putting these two corrections together, if there truly was a brief population bottleneck to a single couple, it is trivial to show that the true
The most straightforward interpretation of this information is that LD, and any other
These corrections had cascading consequences, ultimately undermining almost every claim from population genetics made by Venema and BioLogos. In 2019, Reasons to Believe objected when BioLogos, again, overstated the evidence in population genetics against them.17 But working independently of BioLogos at the time, Stephen Schaffner agreed with RTB’s objection, explaining the corrective,
Estimates of past
are point estimates of the most likely value at each point in time, which is quite a different thing from a lower bound on the population size. also represents a kind of average population size over extended times, rather than a precise estimate for every generation; this makes it rather insensitive to very short bottlenecks.18
One puzzle remains unsolved. The basic fact—that
In 2014, Vern Poythress, a theologian, rightly objected that
LD and
Incomplete Lineage Sorting
The second line of evidence was incomplete lineage sorting (ILS). In the discussion around the book, Venema’s argument from ILS was also quickly dismissed. ILS is another method of computing
ILS uses different data than LD, the genomes of multiple species, not the variation within our species. The approach begins by constructing phylogenetic trees of the across all loci of the genome. In most locations of the genome, humans are closest to chimpanzees, but sometimes there are “discordant” trees, where humans are closer to gorillas. From the details of these discordant trees, we can estimate
The argument looks familiar,
- ILS estimates
as far back as 9 to 18 million years.23 - These estimates of
are always greater than thousands. is the minimum population size consistent with the data. - Therefore, the ILS data demonstrates “large ancestral population sizes throughout the speciation process,”24 back to a time “long before our lineage was even remotely called ‘human.’”25
In the same way as before, this argument is invalid because
The argument from ILS, then, is like, arguing that the US stock market did not crash in 2008, because the average return of the Chinese stock market has always been greater than 10%. Averages do not tell us whether or not the market ever crashed, and the Chinese market is not the US stock market.
ILS is a prominent example of an uncorrected mistake. An article published by the BioLogos organization in 2021 (backdated to 2014) states,
…humans, chimpanzees, and gorillas have, within their genomes, the exact pattern of incomplete lineage sorting predicted by…large ancestral population sizes throughout the speciation process.26
In context, this sentence claims that ILS demonstrates the human lineage had “large ancestral population sizes throughout the speciation process.” But it is also true that the data matches the exact pattern of ILS predicted by a sharp bottleneck at any point in the speciation of humans. ILS does not estimate
Robust Estimates of Minimum Population Size
Since 2021, the BioLogos website states that “Population genetics measures the average population, not a minimum population,” a claim which is just not true.28 To the contrary, there is a broad scientific consensus that population genetics can, indeed, estimate the minimum population size consistent with genetic data, albeit using entirely different methods than effective population size.
At least two lines of evidence robustly estimate the minimum population size, one of which covers the relevant time period: lineage time inference (LiTI) on autosomal DNA, and lineage times of mitochondrial and Y-chromosomal DNA.
Mitochondrial and Y Lineage Times
Though not covering the time period relevant to Venema’s book or revised conclusions, Y-Chromosomal Adam (Y-adam) and Mitochondrial Eve (Mt-eve) set a minimum population size limit for, respectively, men and women. Y-adam is estimated to have lived, roughly, around 150,000 years ago. Mt-eve is estimated to have lived roughly the same time, though they did not likely live at the exact same time and place. This data clearly rules out a single couple bottleneck from Adam and Eve at any time more recent than ~150,000 years ago.
This basic approach to determining minimum population size, lineage time inference, has very broad agreement among scientists, including young earth creationist biologists. For example, Y-adam and Mt-eve feature prominently in Nathaniel Jeanson’s two books on Adam and Eve.29 Jeanson is a biologist with Answers in Genesis, and he reconciles young earth creationism with Y-adam and Mt-eve by using a mutation rate 40x higher than the consensus mutation rate across the genome.30 This mutation rate is the specific point of contention, on which his argument hinges, but this disagreement is framed by a larger point of agreement: lineage times are a valid and understandable approach to estimating minimum population sizes.
Lineage Times of Autosomal DNA
With the collapse of the LD and ILS arguments, an awkward scramble ensued to identify at least some evidence to support the revised conclusion. After rejecting every line of evidence presented by Venema and Schaffner, Buggs proposed the right way forward:31
[compute] the TMR4A for each haplotype block of the human genome… . Until that has been done, I do not think we can say that the bottleneck hypothesis has been rigorously tested.
TMR4A is the “time to most recent 4 alleles,” the time needed for mutations and recombinations to generate, from a single couple, the vast majority of observed human genetic variation across the genome. Compared to LD, ILS, and every other line of evidence considered, Venema agreed,
TMR4A is a better way of looking at this overall—it more directly addresses what we’re really interested in.32
In defense of Venema’s revised conclusions, the TMR4A analysis was implemented and defended almost entirely by another scientist.33
In the end, TMR4A was the primary evidence supporting Venema and BioLogos’s revised conclusions. If the goal is to rule out a single-couple bottleneck, then the most robust and important analysis to present and improve upon are these LiTI estimates of TMR4A.
It had only recently become possible to compute TMR4A across the genome, due to a fascinating advance in population genetics, first was published in 2014. Elegantly accounting for recombination, now several algorithms can infer from genetic variation an “ancestral recombination graph” (ARG), which includes many trees arranged in a sequence along the genome.34 Some ARG inference algorithms are now fast enough to analyze data from thousands of genomes at once.
At each locus of the genome, the ARG’s corresponding tree—from the ideal 4-allele starting point— records the most likely count of mutations and recombinations required to exactly reproduce in fine-grain detail the overwhelming majority of observed human genetic variation.35 The TMR4A date itself is estimated by scaling these counts by the mutation (or recombination) rate across the genome. Each locus of the genome yields a distinct TMR4A, and these estimates across the genome are summarized by their median.
What was the median TMR4A date? For humans alive today, the median TMR4A is approximately 500,000 years ago (Figure 3). Any time more recent, the original couple would need to carry more than four alleles, which is not possible with normal human biology. Richard Buggs, AJ Roberts from RTB, Ann Gauger at the Discovery Institute, and secular scientists at Peaceful Science subjected this analysis to intense and searching scrutiny, publicly raising several objections, but finally agreeing this was a robust result.36
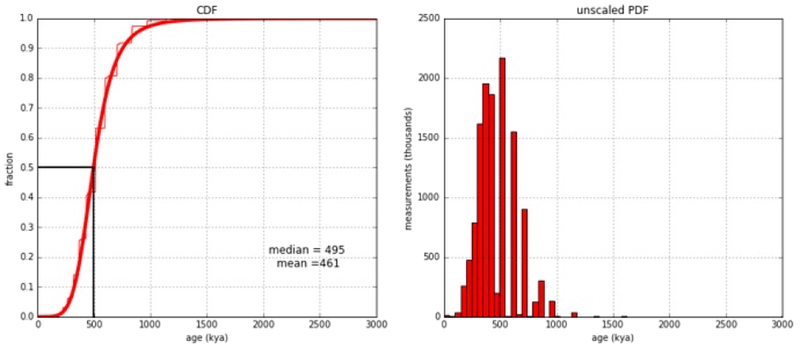
Lineage time inference (LiTI) of TMR4A by this method is both very robust to confounding factors and very resistant to skeptical criticism,37
- TMR4A estimates do not depend on any simulations of demographic history, which are very difficult to accurately execute and are always subject to valid criticism for neglecting important details.38
- The whole autosomal genome is analyzed, neutralizing any objections that the analysis was biased by cherrypicked regions so as to reduce or increase the TMR4A.
- Several factors push estimates lower or higher than the true TMR4A date at any individual locus of the genome, but none of these factors acts across enough of the genome so as to substantially alter the median TMR4A.
- Adding more genomes to the analysis will not decrease the TMR4A date, and empirically new data does not increase the date by much.
- Much as radiometric dating can be crosschecked by comparing independent isotope clocks, TMR4A dates can be computed either using the mutation clock, the recombination clock, or both together.39
A preprint published in 2021 demonstrates that LiTI is very robust indeed, empirically validating TMR4A with several systematic tests.40
There is one caveat to resolve in future work. This TMR4A is computed only from people alive today. Adding in the DNA from archaic humans, such as Neanderthals and Denisovans is more difficult because ancient DNA has many degradation errors that inflate the inferred date. Due to interbreeding in the past, most of the archaic genomes are already included in the current analysis any ways, so the impact of this data may already be baked in. Perhaps TMR4A might increase to 600,000 or 700,000 years ago or stay the same. We do not yet know.
A Broad and Deep Consensus
The consensus around TMR4A is broad and deep. TMR4A features prominently in William Lane Craigs’ response to BioLogos and in his book,41 which was reviewed favorably by secular scientists. Stephen Schaffner, when writing BioLogos’s new position, is silent on the limitations of
RTB used TMR4A and “time to most recent 10 alleles” (TMR10A) to update their model. There were eight people on the Ark: Noah, his wife, his three sons, and their three wives, holding 10 lineages at most.46 Looking across the genome, the median TMR10A is approximately 180,000 years ago. Apart from miraculous intervention or interbreeding between Noah’s lineage and others, the TMR10A rules out a Noahic bottleneck more recent than this time. So RTB places Noah at about 180,000 years ago, and positing that Eve was created as a genetic mosaic, with different genomes in each her eggs, passing on more lineages than needed to account for the LiTI estimates at the origin of our species. Exotic biology, even in context of one or more miracle, will be judged implausible by many. But this model is not in conflict with the genetic evidence.47
Three Disputed Lines of Evidence
There are, additionally, three disputed lines of evidence that were discussed.
Trans-Species Variation
Leaving no stone unturned, we eventually discussed trans-species variation or polymorphisms (TSV or TSP) in detail, coming to a consensus. This line of evidence was not mentioned in Adam and the Genome or any of Venema or BioLogos’s work, TSV appears in the formal scientific literature as a test of a single-couple bottleneck.48 For this reason, it is commonly cited by most biologists when asked questions about Adam and Eve. At Peaceful Science, the discussion continued on with secular scientists for quite some time, eventually arriving to a similar conclusion.
TSP can, at least in principle, place a minimum population limit from present day back to more than 6 million years in the past, long before we became human. Our genome includes person-to-person variation, different versions, or “alleles,” at each locus of the genome. Most of the time, all the variants of a locus within a species are very similar to one another; usually, they are more similar to variants within the species than to variants in other species. In other words, usually the human variants of a gene are all more similar to other human variants than they are to chimpanzee variants.
This pattern is different in a few locations in our genome, usually at loci important for immunity. At these loci, some human variants are much closer to the variants from other species. In other words, in these immune genes, some human variants are closer to chimpanzee variants than they are to other human variants. This pattern of variation is the hallmark of TSV or TSP.
How does TSV arise? How do different species end up with such similar variants? There are two primary mechanisms: (1) shared history of the genes or (2) convergent evolution due to shared selective pressure. Both mechanisms are valid explanations of TSV, and there is dispute about how much is caused by shared history versus convergent evolution. Only TSP caused by shared history places limits on population size, so convergent evolution confounds estimates of minimum population size based on TSP.
How many TSV lineages are observed, and what limits does this place on population size? This is an area of active research, but in the vast majority of loci there are no TSVs. In the few loci where TSV is observed, almost all have less than 4 trans-species lineages.49 In a few cases, such as the HLA genes, there may be up to 10 or 20 shared alleles, though this result has not been replicated. This data, if confirmed and all due to shared history, would place a minimum population limit of 5 to 10 individuals (not thousands) going back 6 million years.
However, HLA genes also show clear evidence of convergent evolution.50 We expect convergent evolution, moreover, in immune genes because different species are often exposed to the same disease-causing organisms. For this reason, TSV-based estimates of minimum population size are lower confidence and disputed.
Simulations of Allele Frequency Spectrum
Allele or Site Frequency Spectrums (AFS or SFS) are a low-information and lossy summary of human genetic variation. This summary is sometimes used in mainstream science to understand population demographics in the somewhat recent past, but the accuracy of AFS inferences degrades rapidly as we look farther back in time. Along with the data underlying LD, the data used by AFS is fully explained and accounted for by the TMR4A estimate.
In the context of the post-publication review of Adam and the Genome, Stephen Schaffner ran several simulations with stable populations of 10,000 individuals arising from a single couple. From these simulations, he computed the AFS and compared it with the AFS observed in African populations (and nowhere else). The observed and simulated AFS did not match until he moved back the single couple origin to about 500,000 years ago. This approach estimates neither a minimum population size nor an effective population size. Rather, it merely tests the specific hypotheses precisely as they are modeled in simulations, in fine-grained detail. Schaffner presented these results to Buggs, who congratulated the attempt, but quickly dismissed the result due to several factors unaccounted for in the simulations.
Eventually, in 2019, Ann Gauger at the Discovery Institute used simulated AFS to compute a date of 500,000 years ago too.51 In 2021 at BioLogos, Schaffner published a different version of the analysis he presented to Buggs, yielding the same date, but using an unnormalized and unscaled AFS.52 At this time, there are at least three reasons this line of evidence is low-confidence and disputed:
- The simulations are dramatically oversimplified in a way that is expected to affect the results, particularly because Schaffner now uses an unnormalized AFS.53
- Creationists published simulations with a much more recent Adam and Eve and different demographic details, generating an AFS matching the observed data. Schaffner does not cite or explain what is invalid about this body of work.54
- I was unable to replicate Schaffner’s results using simulation software commonly used in the field. His write up was exceedingly terse, without enough information to clarify the source of the discrepancy. Schaffner was non-responsive to questions.
It is possible that future work might develop AFS into a useful line of evidence. The simulations on which the analysis is based need to match both the relevant models of Adam and Eve and the known details of demographic history, also controlling for sampling effects, and systematically determining the starting genotype of the original couple. This sort of modeling is very difficult to do with confidence, and AFS inferences based on simulations will always be vulnerable to criticism for neglecting important details.
Interbreeding and Admixing
There is very good evidence of interbreeding—sometimes referred to as “admixing” or “admixture”—between Homo sapiens, Neanderthals and Denisovans, but the interpretation of this interbreeding is disputed.
Strictly speaking, interbreeding does rule out the genetic-bottleneck model of Venema’s revised claims. But Adam and the Genome, and the BioLogos organization, made much bigger claims than admixing evidence supports, which is why the book itself just notes interbreeding in passing. For the last decade or more, essentially every creationist organization agreed that Homo sapiens and Neandertals interbred.55 If this admixing were all that was claimed, there would have been no controversy.
Most critically important, interbreeding between Homo sapiens and Neanderthals disconnects the evidence from claims made by BioLogos more recently. Were there ever just two Homo sapiens?56 As was made clear in post-publication review, this question has nothing to do with whether there was a genetic bottleneck down to a single couple in our lineage.
In late 2019, Jeff Schloss, a biologist associated with BioLogos, submitted an article explaining that population sizes estimates do not have “taxonomic specificity.”57 There is no plausible way to sort out how many of our ancestors at the relevant times are Homo sapiens or not. Consequently, large “ancestral” population size cannot, even in principle, demonstrate there was never just two Homo sapiens.
BioLogos declined to publish this article, in favor of publishing a more diffuse statement of error in early 2020. Ignoring Schloss’s correction, this statement went on to present large average ancestral population sizes as evidence that there was never “just two Homo sapiens.”58
Zombie Science and Defunct Citations
For three reasons, the faith-science conversation may be uniquely susceptible to zombie science from population genetics. First, several false claims based on population genetics are theologically potent, and the change to the consensus is not yet widely known. Second, most scholars in this space do not have the expertise to evaluate claims from population genetics, which are technical and frequently over-extended. Third, quite a bit of scholarship is published in blogs, books, and other venues with inadequate or non-existent peer review.
If we are to discuss genetic bottlenecks, the scientific claims we make about them should be accurate and grounded in good science. And valid claims about bottlenecks should not be over-extended to make negative claims about more sensible models of Adam and Eve, or the theology associated with these models.
Examples of Zombie Science
In 2021, Perspectives on Science and the Christian Faith (PSCF) published a series of three articles examining an approach to original sin that does not require descent from Adam and Eve.59 Motivating their project, however, the series presumes that the scientific evidence conflicts with universal descent from Adam and Eve, authoritatively citing a defunct article, known since 2018 to be in error.60 The series could have still made their theological case, but on a level playing field, without appeals to defunct articles.
A recent book by Loren Haarsma, When Did Sin Begin?, repeats several incorrect scientific claims. On the one hand, this book’s summary of the evidence presents a very different position than BioLogos held in the past. Recent universal ancestry is presented early on.61 Viable models of de novo creation of Adam and Eve are presented, and the book makes clear that the “humans” of theology need not correspond to specific taxonomic categories, such as Homo sapiens.62
On the other hand, the book makes several defunct scientific claims, referencing several defunct citations in support of these claims, never once engaging corrective scholarship. Most prominently, the book authoritatively references Adam and the Genome, without citing or discussing the post-publication review that corrected the book.
The book authoritatively cites a deleted article at BioLogos,63 claiming LD and ILS, might, after all, rule out a single-couple bottleneck going back millions of years. In a material omission, there is no explanation that estimates of
Without engaging or citing corrective scholarship, the book repeats BioLogos’s revised argument, that
These claims are incorrect and cannot be understood as informed scientific disagreement. This book does not present a new scientific argument, nor does it cite or discuss the scholarship that corrects its references. Without engaging corrective scholarship or acknowledging the corrections themselves, this book resurrects zombie science that was vanquished long ago by peer-review.
Neither Complete nor Transparent
The academic discourse on faith and science is interdisciplinary; we rely on one another to accurately represent the findings of our respective fields.
The BioLogos organization, by 2021, deleted or substantially revised at least 9 articles on their website.64 The article that first brought their scientific argument into the public eye was deleted.65 Every article supporting the Christianity Today coverage of BioLogos’s scientific position was deleted.66 Venema’s articles responding to Buggs were also deleted.67 One deleted article was substantially revised, backdated to 2014, and reposted under new authorship and with altered conclusions.68 This solitary article is now the lone defense of the coverage by Christianity Today in 2011. The revisions removed discussion of LD, but still present ILS as a relevant line of evidence.
BioLogos is a well-respected organization that has done a lot of good, credible in large part due to its founder and former NIH-director, Francis Collins. When asked to make transparent corrections, BioLogos declined, explaining the organization’s website is merely a blog.69 The organization does not follow the baseline publication ethics of an academic journal, a research organization, or even a journalistic outlet.
Because they are a credible organization, this response creates a confusion situation, in which defunct citations and scientific errors are propagating.
Conclusions
This review explains what population genetics tells us about single-couple genetic bottlenecks. Special attention was given to explaining each line of evidence, and how our understanding has changed over the last decade.
Acknowledgement
This scholarship was supported by a visiting professorship funded by the John Templeton Foundation and hosted by the Southeastern Baptist Theological Seminary in 2021-2022. The author is grateful to several scholars for reading and providing feedback on a draft of this manuscript: John Wood, Ken Keathley, Michael Murray, William Lane Craig, Ken Turner, Andrew Loke, Jeff Hardin, Jeff Schloss, Greg Cootsona, Gerry McDermott, AJ Roberts, Glenn Branch, Ted Davis, and Daniel Ang.
Deborah Haarsma, Jim Stump, Loren Haarsma, Ard Louis, and Dennis Venema from BioLogos were also presented a draft of this manuscript, but they did not provide a response.
References
Ayala, F J, a Escalante, C O’Huigin, and J Klein, ‘Molecular Genetics of Speciation and Human Origins.’, Proceedings of the National Academy of Sciences of the United States of America, 91.15 (1994), 6787–94 https://doi.org/10.1073/pnas.91.15.6787.
Ayala, Francisco J., and Ananias A. Escalante, ‘The Evolution of Human Populations: A Molecular Perspective’, Molecular Phylogenetics and Evolution, 5.1 (1996), 188–201 https://doi.org/10.1006/mpev.1996.0013.
BioLogos Editorial Team, ‘Adam, Eve, and Human Population Genetics’, BioLogos, 2021.
BioLogos Forum, ‘Forum Discussion on “Adam, Eve and Population Genetics: A Reply to Dr. Richard Buggs (Part 1)”’, BioLogos, 2018 https://doi.org/10.5281/zenodo.1323939.
Bradley Hagerty, Barbara, ‘Evangelicals Question The Existence Of Adam And Eve’, National Public Radio, 2011 [accessed 10 October 2020].
Buggs, Richard, ‘Adam and Eve: A Tested Hypothesis?’, Nature Portfolio Ecology & Evolution Community, 2017 [accessed 6 November 2021].
Buggs, Richard, ‘Adam and Eve: Lessons Learned’ [accessed 8 July 2022].
Clouser, Roy, ‘Three Theological Arguments in Support of Carol Hill’s Reading of the Historicity of Genesis and Original Sin’, Perspectives on Science and Christian Faith, 73.3 (2021), 7.
Craig, William Lane, ‘What Became of the Genetic Challenge to Adam and Eve?’, Peaceful Science, 2020 https://doi.org/10.54739/e7r9.
Elliot-Higgins, Jack, and S. Joshua Swamidass, ‘Brief Population Bottlenecks Are Beyond The Genetic Streetlight’, 2021 https://doi.org/10.21203/rs.3.rs-817056/v1.
Haarsma, Deborah, ‘Truth-Seeking in Science’, BioLogos, 2020 [accessed 3 July 2022].
Haarsma, Loren, ‘When Did Sin Begin?’ (Baker Academic, 2021).
Hill, Carol A, ‘Original Sin with Respect to Science, Origins, Historicity of Genesis, and Traditional Church Views’, Perspectives on Science and Christian Faith, 73.3 (2021), 14.
Hobolth, Asger, Julien Y. Dutheil, John Hawks, Mikkel H. Schierup, and Thomas Mailund, ‘Incomplete Lineage Sorting Patterns among Human, Chimpanzee, and Orangutan Suggest Recent Orangutan Speciation and Widespread Selection’, Genome Research, 21.3 (2011), 349–56 https://doi.org/10.1101/gr.114751.110.
Horst, William, ‘From One Person? Exegetical Alternatives to a Monogenetic Reading of Acts 17:26’, Perspectives on Science and Christian Faith, 74.2 (2022), 15.
Hossjer, Ola, and Ann Gauger, ‘A Single-Couple Human Origin Is Possible’, BIO-Complexity, 2019.1 (2019) https://doi.org/10.5048/BIO-C.2019.1.
Jeanson, Nathaniel, ‘Evidence for a Human Y Chromosome Molecular Clock’, Answers Research Journal, 2019 [accessed 9 July 2022].
Jeanson, Nathaniel, Replacing Darwin: The New Origin of Species (Master Books, 2017).
Jeanson, Nathaniel, Traced: Human DNA’s Big Surprise (Master Books, 2022).
Kemp, Kenneth W, ‘Science, Theology, and Monogenesis’, American Catholic Philosophical Quarterly, 85.2 (2011), 217–36 https://doi.org/10.5840/acpq201185213.
Leffler, Ellen M., Ziyue Gao, Susanne Pfeifer, Laure Ségurel, Adam Auton, Oliver Venn, Rory Bowden, Ronald Bontrop, Jeffrey D. Wall, Guy Sella, Peter Donnelly, Gilean McVean, and Molly Przeworski, ‘Multiple Instances of Ancient Balancing Selection Shared Between Humans and Chimpanzees’ [accessed 8 July 2022] https://www.science.org/doi/10.1126/science.1234070.
Loke, Andrew Ter Ern, ‘Reconciling Evolution and Biblical Literalism: A Proposed Research Program’, Theology and Science, 14.2 (2016), 160–74 https://doi.org/10.1080/14746700.2016.1156328.
McEvoy, Brian P., Joseph E. Powell, Michael E. Goddard, and Peter M. Visscher, ‘Human Population Dispersal “Out of Africa” Estimated from Linkage Disequilibrium and Allele Frequencies of SNPs’, Genome Research, 21.6 (2011), 821–29 https://doi.org/10.1101/gr.119636.110.
McKnight, Scott and Dennis R. Venema, Adam and the Genome: Reading Scripture after Genetic Science, 2017.
Murphy, George L, ‘The Twofold Character of Original Sin in the Real World’, Perspectives on Science and Christian Faith, 73.3 (2021), 6.
Ostling, Richard N., ‘The Search for the Historical Adam’, Christianity Today, 2011.
Peaceful Science Forum, ‘Rob Carter Responds to TMR4A’, Peaceful Science, 2018 [accessed 8 July 2022].
Poythress, Vern S., Did Adam Exist? (P & R Publishing, 2014).
R. Vahdati, Ali, and Andreas Wagner, ‘Parallel or Convergent Evolution in Human Population Genomic Data Revealed by Genotype Networks’, BMC Evolutionary Biology, 16.1 (2016), 154 https://doi.org/10.1186/s12862-016-0722-0.
Roberts, Anjeanette, ‘How Can Christians Disagree over Adam and Eve?’, Reasons to Believe, 2019 [accessed 22 July 2022].
Roberts, Anjeanette, ‘Mosaic Eve: Mother of All (Part 1)’, Reasons to Believe, 2020 [accessed 5 July 2022].
Roberts, Anjeanette, ‘Mosaic Eve: Mother of All (Part 2)’, Reasons to Believe, 2020 [accessed 5 July 2022].
Sanford, John, Robert Carter, Wes Brewer, John Baumgardner, Bruce Potter, and Jon Potter, ‘Adam and Eve, Designed Diversity, and Allele Frequencies’, Proceedings of the International Conference on Creationism, 8.1 (2018) https://doi.org/10.15385/jpicc.2018.8.1.20.
Scally, Aylwyn, ‘The Mutation Rate in Human Evolution and Demographic Inference’, Current Opinion in Genetics & Development, 41 (2016), 36–43 https://doi.org/10.1016/j.gde.2016.07.008.
Schaffner, Stephen, ‘What Genetics Says About Adam and Eve’ [accessed 8 July 2022]
Swamidass, S. Joshua, ‘A U-Turn on Adam and Eve’, Peaceful Science, 2021 https://doi.org/10.54739/83rj.
Swamidass, S. Joshua, ‘BioLogos Deletes an Article’ https://doi.org/10.54739/rv8k, Peaceful Science, 2021 https://doi.org/10.54739/83rj.
Swamidass, S. Joshua, ‘Heliocentric Certainty Against a Bottleneck of Two?’, 2018 https://doi.org/10.5281/zenodo.1328247.
Swamidass, S. Joshua, The Genealogical Adam and Eve: The Surprising Science of Universal Ancestry (InterVarsity Press, 2019).
Swamidass, S. Joshua, ‘The Overlooked Science of Genealogical Ancestry’, Perspectives on Science and Christian Faith, 70.1 (2018).
Tenesa, Albert, Pau Navarro, Ben J. Hayes, David L. Duffy, Geraldine M. Clarke, Mike E. Goddard, and others, ‘Recent Human Effective Population Size Estimated from Linkage Disequilibrium’, Genome Research, 17.4 (2007), 520–26 https://doi.org/10.1101/gr.6023607.
Venema, Dennis R., ‘Adam, Eve and Human Population Genetics’, BioLogos, 2014 https://doi.org/10.54739/d67o.
Venema, Dennis R., ‘Adam, Eve and Population Genetics: A Reply to Dr. Richard Buggs (Part 1)’, BioLogos, 2017 https://doi.org/10.54739/ju3t.
Venema, Dennis R., ‘Adam, Eve and Population Genetics: A Reply to Dr. Richard Buggs (Part 2)’, BioLogos, 2017 https://doi.org/10.54739/2xqh.
Venema, Dennis R., ‘Genesis and the Genome: Genomics Evidence for Human-Ape Common Ancestry and Ancestral Hominid Population Sizes’, Perspectives on Science and Christian Faith, 62.3 (2010).
Venema, Dennis R., ‘Vern Poythress, Population Genomics, and Locating the Historical Adam’, BioLogos, 2015 https://doi.org/10.54739/atjo.
Venema, Dennis R., and Darrel R. Falk, ‘Does Genetics Point to a Single Primal Couple?’, BioLogos, 2010 https://doi.org/10.54739/vqbj.
Mar 20, 2023
Mar 23, 2023
Jul 5, 2025


